Snakemake is a workflow management tool implemented as extra magic on top of Python. If you want to take your Snakemake workflows from beginner to advanced, learning a little Python goes a long way.
This is just enough Python to understand and apply some cool tricks in your Snakemake workflows.
prereqs
- Basic Snakemake knowledge. You’ve gone through the official tutorial. Maybe you’ve even built your own snakemake workflow.
- Minimal or no python knowledge, aside from what you may have gleaned from the snakemake docs.
- Basic R knowledge. Tidyverse experience is helpful but not required.
variables
Here are the basic variable types in Python and the equivalent type in R:
basic types
| Python | R |
|---|---|
| string | character |
| integer | integer |
| float | numeric |
| boolean | logical |
| None | NULL |
A key difference is that in R, all of the above objects are vectors of length 1.
type casting
You can convert variables to different types:
The equivalent R code:
[1] 32[1] 32[1] "32"[1] FALSE[1] TRUElogical(0)What similarities and differences do you notice when type-casting in Python vs. R?
Naming variables
The biggest difference in variable names between R and Python: no dots are allowed in Python variable names. All other rules from R apply. Python variable names must start with an alphabetic character and they can contain alphanumeric characters and underscores.
Just like in R, it is a good idea not to name a variable the same thing as a reserved keyword or global function.
Here is a list of the Python keywords. Enter any keyword to get more help.
False break for not
None class from or
True continue global pass
__peg_parser__ def if raise
and del import return
as elif in try
assert else is while
async except lambda with
await finally nonlocal yieldformatting strings
You can add strings together to build new ones like this:
Python
Madame Marie S. CurieThe print function takes any number of arguments and will insert a space between items by default:
But there’s a better way!
f-Strings in Python work just like glue() in R, where curly braces in the string are interpreted as code. They’re much easier to read & write:
This is equivalent to:
container types
Here are a few of the most useful container types and their equivalents in R:
| Python | R |
|---|---|
| list | vector (all same type) or list (multiple types) |
| dictionary | named list |
| DataFrame (pandas) | data.frame (base) or tibble (tidyverse) |
lists
Python lists are most similar to lists in R. Python lists can also be compared to vectors in R, but unlike vectors, lists can contain objects of different types.
Python
['apple', 'banana', 'grape', 'pizza', 'fries', 3, -2.5, 'donuts']['apple', 'banana', 'grape', 'mango']Indexing
Indexing is 0-based in Python. You can index lists and strings.
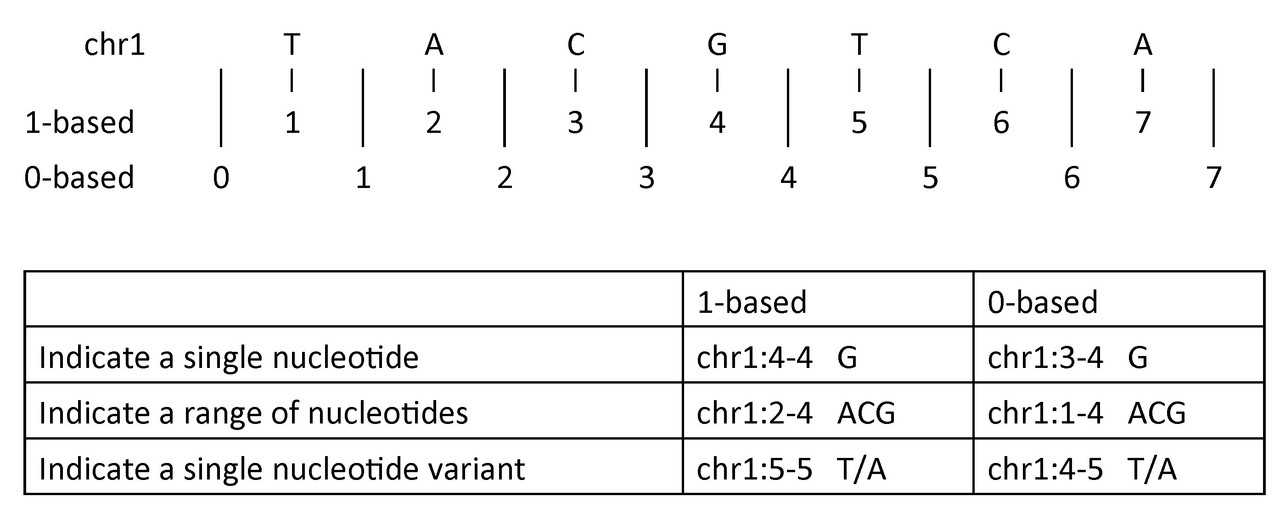
4144You might find 0-based indexing unintuitive if you’re used to working in languages like R that use 1-based indexing, but this is “not mad, just differently sane!”2
Here’s an illustration that shows how 0-based & 1-based indexing systems compare in the context of DNA coordinates3:
We can try to write equivalent code in R…
But this doesn’t work! In the above R code, dna is a vector of length 1 with “TACGTCA” as the item at index 1. You can’t grab individual characters out of it. We have to break up the string into a vector with every character as a separate item in R.
dictionaries
Dictionaries consist of key-value pairs where you use the key to access the value. Keys can be strings, integers, floats, or booleans. Values can be anything!
'cherry'['carrot', 'eggplant']'carrot'Dictionaries can be nested:
Python
0.5Get a list of keys in the dictionary with:
Snakemake config
Say you have a Snakemake workflow for training machine learning models. You want to be able to specify different datasets, ML methods, and random seeds, so you use f-Strings to fill in these variables:
You can improve this by creating a configuration file in YAML format:
Then specify your config file at the top of your Snakefile. Snakemake parses your YAML config files to a dictionary called config.
When you run this workflow, it uses config/otu.yml by default:
rule train_ml_model:
input: workflow/scripts/train_ml.R, data/OTU.csv
output: results/OTU/runs/rf_1001_model.Rds
jobid: 0
reason: Missing output files: results/OTU/runs/rf_1001_model.Rds
resources: tmpdir=/var/folders/9n/glrhxtfx453gl68sc1gzq0mc0000gr/T
Job stats:
job count min threads max threads
-------------- ------- ------------- -------------
train_ml_model 1 1 1
total 1 1 1Now you can change the values of these variables by editing the config file, or by creating multiple config files and specifying them on the command line:
rule train_ml_model:
input: workflow/scripts/train_ml.R, data/genus.csv
output: results/genus/runs/glmnet_1001_model.Rds
jobid: 0
reason: Missing output files: results/genus/runs/glmnet_1001_model.Rds
resources: tmpdir=/var/folders/9n/glrhxtfx453gl68sc1gzq0mc0000gr/T
Job stats:
job count min threads max threads
-------------- ------- ------------- -------------
train_ml_model 1 1 1
total 1 1 1See this example in context here.
conditionals
Think of these as functions that return a boolean.
| operator | meaning | same in R |
|---|---|---|
== |
equal | ✅ |
!= |
not equal | ✅ |
< |
less than | ✅ |
> |
greater than | ✅ |
<= |
less than or equal to | ✅ |
>= |
greater than or equal to | ✅ |
| or | or (inclusive) | | |
| and | and | & |
| in | in | %in% |
| not | not | ! |
if-else statements
No curly braces here! But indentation does matter.
Python
This list is shortinline if-else statement
When you create a variable with an if-else statement, you can do it all on one line:
This is equivalent to:
You can’t make it a one-liner if you need to use elif.
nested if-else blocks
for loops
If a variable has a length (i.e. you can run len(myvar)), then you can iterate over it in a for loop.
Python
Allison Mason, PhD
Courtney Armour, PhD
Sarah Lucas, PhDrange
Use range() to loop over a range of numbers. If you give range() two arguments, range starts counting from the first argument and stops just before the second argument, stepping by 1. In other words, the stop argument is exclusive.
1 bottles of pop on the wall
2 bottles of pop on the wall
3 bottles of pop on the wallIf you give range just one argument, it starts counting from zero and uses the argument as the stop value:
0 bottles of pop on the wall
1 bottles of pop on the wall
2 bottles of pop on the wall
3 bottles of pop on the wallYou can also use range() to create lists:
You can use range with three arguments: start, stop, and step:
Give the step argument a negative value and make start greater than stop to move backwards:
You might think it’s weird that range stops just before the stop value. But it works nicely when you give it the length of the list – now you have the list indices!
iterate over dictionaries
Python
There are only 10 hard problems in computer science: 1 naming things
2 understanding binary
3 off-by-one errorslist comprehensions
When you want to make a for loop that creates a new list, dictionary, or set, use a comprehension. I’m only going to cover list comprehensions below, but you can learn about dictionary and set comprehensions here.
To build a list with a for loop, you can do this:
but a list comprehension is much sleeker and performs faster:
This is roughly equivalent to using *apply functions in R:
You can continue nesting for x in y statements to build complex lists like this:
['123', '124', '133', '134', '223', '224', '233', '234']Snakemake expand
Let’s say you have a Snakemake workflow with a rule that should run multiple times with different parameters. For example, you may want to train machine learning models with different ML methods, datasets, and random seeds. You define these as wildcards in a rule to train each ML model:
Notice that we’re not using f-Strings within the rule because we want Snakemake to recognize these as wildcards and fill in different values for them with each run of the train_ml_model rule.
To tell Snakemake what values to assign for these wildcards, you can create lists with the values you want and write a target rule at the top of your Snakefile.
Here, you could use an f-String and a list comprehension to create the list of target files.
Python
['results/OTU/runs/rf_1000_model.Rds', 'results/OTU/runs/rf_1001_model.Rds', 'results/OTU/runs/rf_1002_model.Rds', 'results/OTU/runs/rf_1003_model.Rds', 'results/OTU/runs/rf_1004_model.Rds', 'results/OTU/runs/glmnet_1000_model.Rds', 'results/OTU/runs/glmnet_1001_model.Rds', 'results/OTU/runs/glmnet_1002_model.Rds', 'results/OTU/runs/glmnet_1003_model.Rds', 'results/OTU/runs/glmnet_1004_model.Rds', 'results/OTU/runs/svmRadial_1000_model.Rds', 'results/OTU/runs/svmRadial_1001_model.Rds', 'results/OTU/runs/svmRadial_1002_model.Rds', 'results/OTU/runs/svmRadial_1003_model.Rds', 'results/OTU/runs/svmRadial_1004_model.Rds', 'results/genus/runs/rf_1000_model.Rds', 'results/genus/runs/rf_1001_model.Rds', 'results/genus/runs/rf_1002_model.Rds', 'results/genus/runs/rf_1003_model.Rds', 'results/genus/runs/rf_1004_model.Rds', 'results/genus/runs/glmnet_1000_model.Rds', 'results/genus/runs/glmnet_1001_model.Rds', 'results/genus/runs/glmnet_1002_model.Rds', 'results/genus/runs/glmnet_1003_model.Rds', 'results/genus/runs/glmnet_1004_model.Rds', 'results/genus/runs/svmRadial_1000_model.Rds', 'results/genus/runs/svmRadial_1001_model.Rds', 'results/genus/runs/svmRadial_1002_model.Rds', 'results/genus/runs/svmRadial_1003_model.Rds', 'results/genus/runs/svmRadial_1004_model.Rds']Here’s what your Snakefile looks like now.
Snakefile
datasets = ['OTU', 'genus']
methods = ['rf', 'glmnet', 'svmRadial']
seeds = range(1000, 1005)
rule targets:
input:
[f"results/{dataset}/runs/{method}_{seed}_model.Rds" for dataset in datasets for method in methods for seed in seeds]
rule train_ml_model:
input:
R="workflow/scripts/train_ml.R",
csv=f"data/{dataset}.csv"
output:
model="results/{dataset}/runs/{method}_{seed}_model.Rds"
script:
"scripts/train_ml.R"The list comprehension gets harder to read the more for statements you add to it. So Snakemake provides a function expand() to clean this up:
Snakefile
datasets = ['OTU', 'genus']
methods = ['rf', 'glmnet', 'svmRadial']
seeds = range(1000, 1005)
rule targets:
input:
expand("results/{dataset}/runs/{method}_{seed}_model.Rds",
dataset = datasets, method = methods, seed = seeds)
rule train_ml_model:
input:
R="workflow/scripts/train_ml.R"
output:
model="results/{dataset}/runs/{method}_{seed}_model.Rds"
script:
"scripts/train_ml.R"Using expand() creates the exact same list as the list comprehension, but it’s much easier to read and add variables to.
functions
You can define your own functions in R:
and in Python:
anonymous functions
Sometimes you want to write a simple function that you only use once. They’re so inconsequential you don’t even want to give them a name.
We’ve already used one in R4 inside sapply() above:
In Python you use the lambda keyword:
Before the colon, list the arguments of the function. After the colon, compute the value that the function should return.
lambda in Snakemake
When writing Snakemake workflows, lambda functions are useful for defining input files based on the wildcards in output files. Consider a workflow where you have several rules that plot figures for a manuscript. When you initially conduct the analysis, you don’t know how the figures will be ordered in the manuscript. Once you begin drafting the manuscript, you decide that your diversity plot will be figure 1 and your error rates plot will be figure 2. You also decide to convert the figures to a different format, so the conversion step seems like a good opportunity to rename the figures. Initially you write this workflow:
Snakefile
rule targets:
input:
"paper/paper.pdf"
rule convert_figure_1:
input:
tiff='figures/diversity.tiff'
output:
png="paper/figure_1.png"
shell:
"""
convert {input.tiff} {output.png}
"""
rule convert_figure_2:
input:
tiff='figures/error_rates.tiff'
output:
png="paper/figure_2.png"
shell:
"""
convert {input.tiff} {output.png}
"""
rule render_paper:
input:
Rmd="paper/paper.Rmd",
R="workflow/scripts/render_rmd.R",
figures=['paper/figure_1.png', 'paper/figure_2.png']
output:
pdf="paper/paper.pdf"
script:
"scripts/render_rmd.R"The rules convert_figure_1 and convert_figure_2 are a bit repetitive; they only differ by the input filenames and the figure numbers in the output filenames. Maybe this isn’t so bad with only two figures, but you might actually have 5-10 figures for a full scientific paper. We can reduce the repetitive code with a few tricks:
- Create a dictionary
figures_dictthat maps the figure numbers to the descriptive figure file names. - Use a single rule to convert figures called
convert_tiff_to_png, using a lambda function to get the input figure filenames based on the final figure numbers.
Snakefile
figures_dict = {'1': 'diversity', '2': 'error_rates'}
rule targets:
input:
"paper/paper.pdf"
rule convert_tiff_to_png:
input:
tiff=lambda wildcards: f"figures/{figures_dict[wildcards.fig_num]}.tiff"
output:
png="paper/figure_{fig_num}.png"
shell:
"""
convert {input.tiff} {output.png}
"""
rule render_paper:
input:
Rmd="paper/paper.Rmd",
R="workflow/scripts/render_rmd.R",
figures=['paper/figure_1.png', 'paper/figure_2.png']
output:
pdf="paper/paper.pdf"
script:
"scripts/render_rmd.R"This lambda function is equivalent to:
This works because Snakemake allows you to define a function that takes the rule’s wildcards and returns a list of input filenames, rather than literally listing the input filenames as before. This greatly reduces the repetitiveness of the code and makes it easier to maintain.
We can improve this Snakefile even further by replacing the list of figures in render_paper with a call to expand():
Snakefile
figures_dict = {'1': 'diversity', '2': 'error_rates'}
rule targets:
input:
"paper/paper.pdf"
rule convert_tiff_to_png:
input:
tiff=lambda wildcards: f"figures/{figures_dict[wildcards.fig_num]}.tiff"
output:
png="paper/figure_{fig_num}.png"
shell:
"""
convert {input.tiff} {output.png}
"""
rule render_paper:
input:
Rmd="paper/paper.Rmd",
R="workflow/scripts/render_rmd.R",
figures=expand(rules.convert_tiff_to_png.output.png,
fig_num = figures_dict.keys())
output:
pdf="paper/paper.pdf"
script:
"scripts/render_rmd.R"You can take a look at this full example in context here. Also see the Snakemake docs for more about input functions and params functions.
recap
Snakemake concepts covered
- Use f-Strings for human-readable string formatting.
- Config files are imported as dictionaries and allow you to change workflow parameters without modifying code.
- Snakemake’s expand() function is a readable way to build lists just like list comprehensions.
- Lambda functions help define Snakemake input files based on wildcards. You can also use them to define params based on wildcards and/or output files.
resources
Footnotes
The Python logo by the Python Software Foundation is licensed under GNU GPL v2. The Snakemake logo by Johannes Köster is licensed under CC BY-SA 4.0. The R logo by Hadley Wickham and others at RStudio is licensed under CC BY-SA 4.0.↩︎
Greg Wilson quoting Terry Pratchett in The Tidynomicon: https://tidynomicon.github.io/tidynomicon/↩︎
from biostars: https://www.biostars.org/p/84686/↩︎
Read more about anonymous functions in Hadley Wickham’s book Advanced R: http://adv-r.had.co.nz/Functional-programming.html#anonymous-functions↩︎